The combination of Shimadzu microchip technology and automated analysis technology enables analysis that is cheaper,faster,more sensitive and more accurate than agarose gel elecrtrophoresis analysis.
A sophisticated reusable microchip achieves lower running costs for consumables per analysis than agarose gel electrophoresis.
Reusable microchips and selecting the optimal reagent for each sample achieves excellent analytical performance. High-speed automatic operation for up to 120 analyses (An analysis schedule can include up to 120 samples while the instrument can accept a maximum of 108 samples (96 + 12 extra).) - Approx. 80 seconds/analysis processing speed
This instrument is equipped with a LED-excited fluorescence detector. It's more than 10 times as sensitive as ethidium bromide staining.
Select the optimal separation buffer that suits the sample. Simultaneous electrophoresis of an internal marker ensures high analysis reliability and reproducibility.
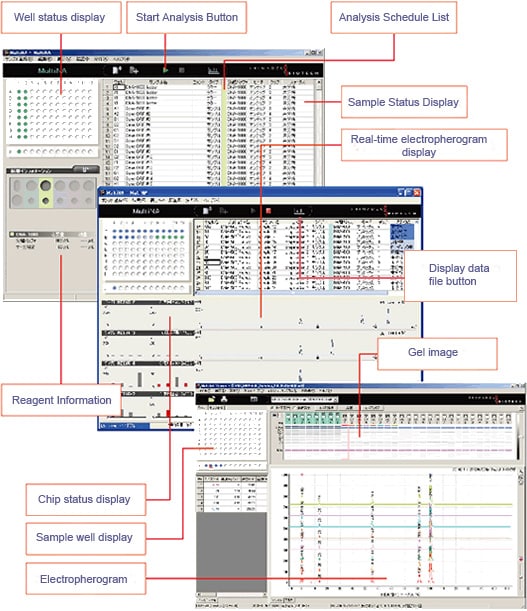
Extremely simple operation. Once the analysis schedule has been created, simply load the samples and reagents and click the Start button.
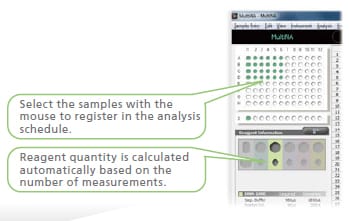
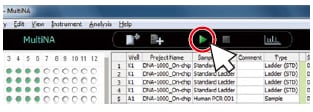
1. Register the analysis schedule.*
* A single analysis schedule permits analysis using multiple reagent kits.
2.Load the samples and reagents.

3. Press the Start button.
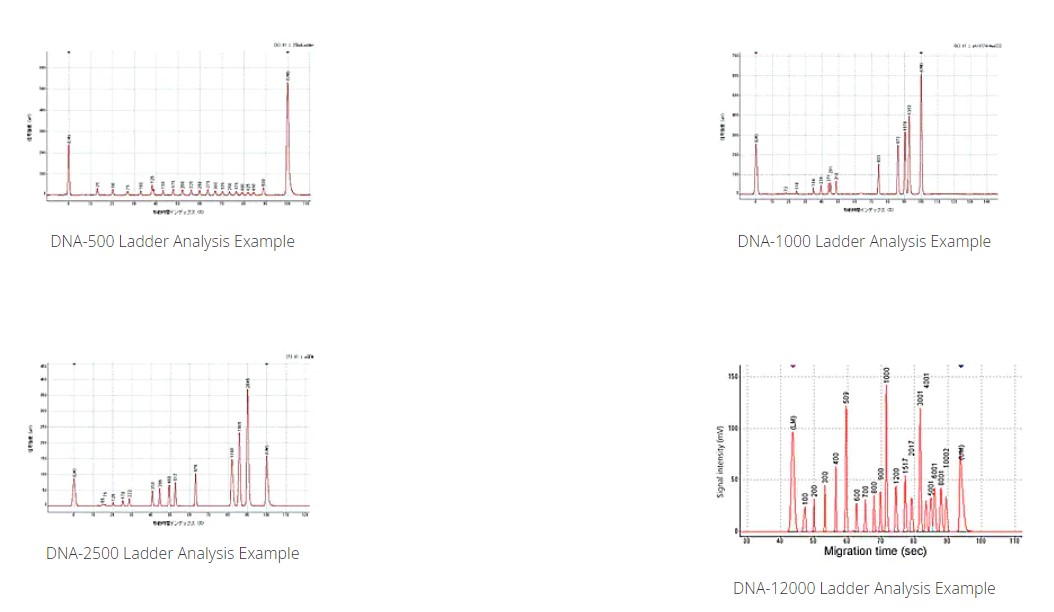
Shimadzu's BioMEMS microchip technology not only enables high-speed analysis, but also achieves high separation performance and reproducibility. Our newly developed separation buffer, analytical method and automation technology overcome the shortness of the effective separation length. Shown below are examples that demonstrate the enhanced analytical performance.
Analysis Using DNA-500/1000/2500/12000 Kit Electrophoretic separation correction and generation of the size calibration curve are conducted automatically with the ladder and 2 internal standard markers (LM and UM in the figure). *Red: DNA ladder